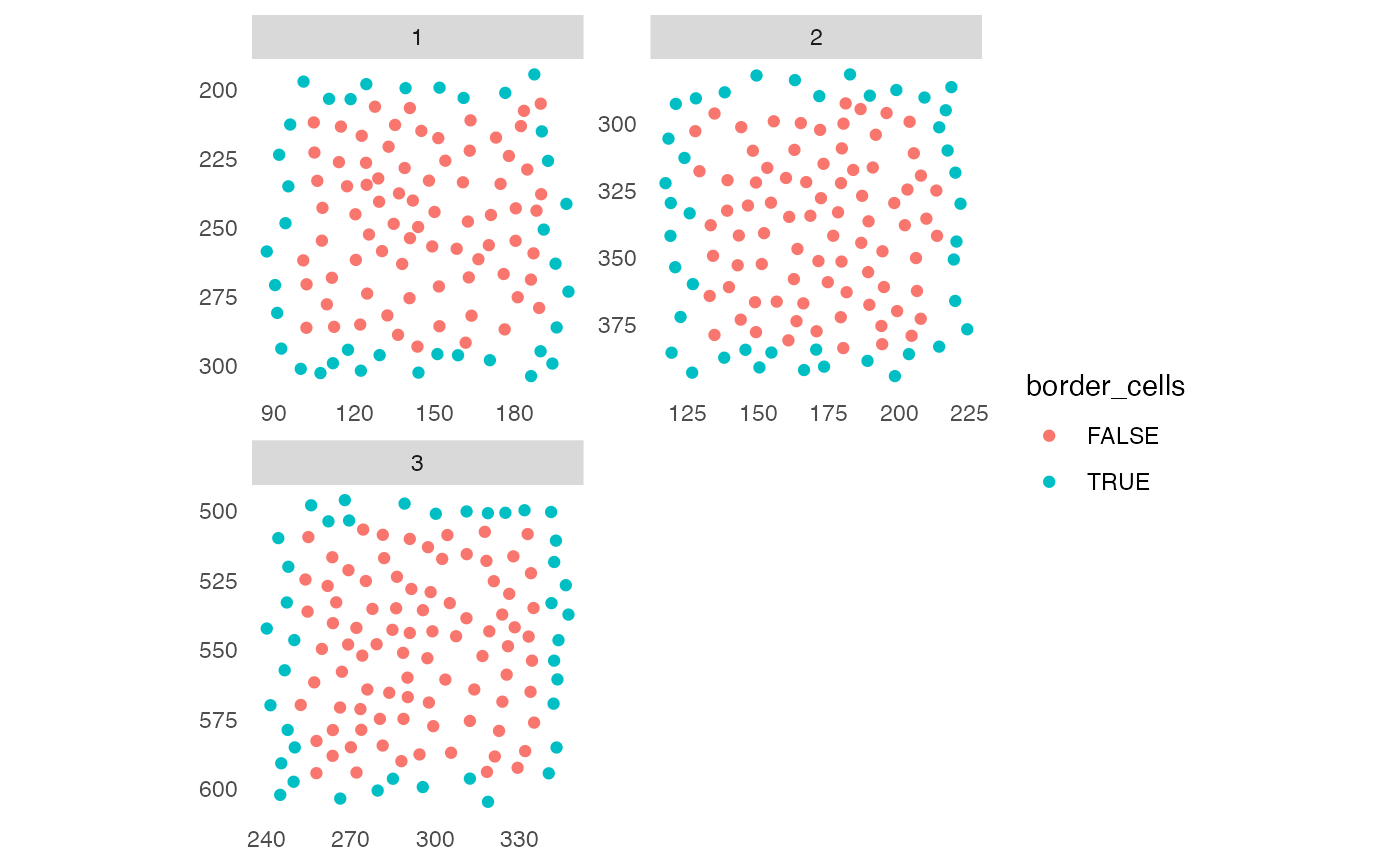
Detection of cells close to the image border for subsequent exclusion from downstream analyses.
findBorderCells(object, img_id, border_dist, coords = c("Pos_X", "Pos_Y"))Arguments
- object
a
SingleCellExperimentorSpatialExperimentobject.- img_id
single character indicating the
colData(object)entry containing the unique image identifiers.- border_dist
single numeric defining the distance to the image border. The image border here is defined as the minimum and maximum among the cells' x and y location.
- coords
character vector of length 2 specifying the names of the
colData(for aSingleCellExperimentobject) or thespatialCoordsentries indicating the cells' x and y locations.
Value
an object of class(object) containing the logical
border_cells entry in the colData slot.
Examples
library(cytomapper)
data("pancreasSCE")
sce <- findBorderCells(pancreasSCE, img_id = "ImageNb",
border_dist = 10)
plotSpatial(sce,
img_id = "ImageNb",
node_color_by = "border_cells",
scales = "free")