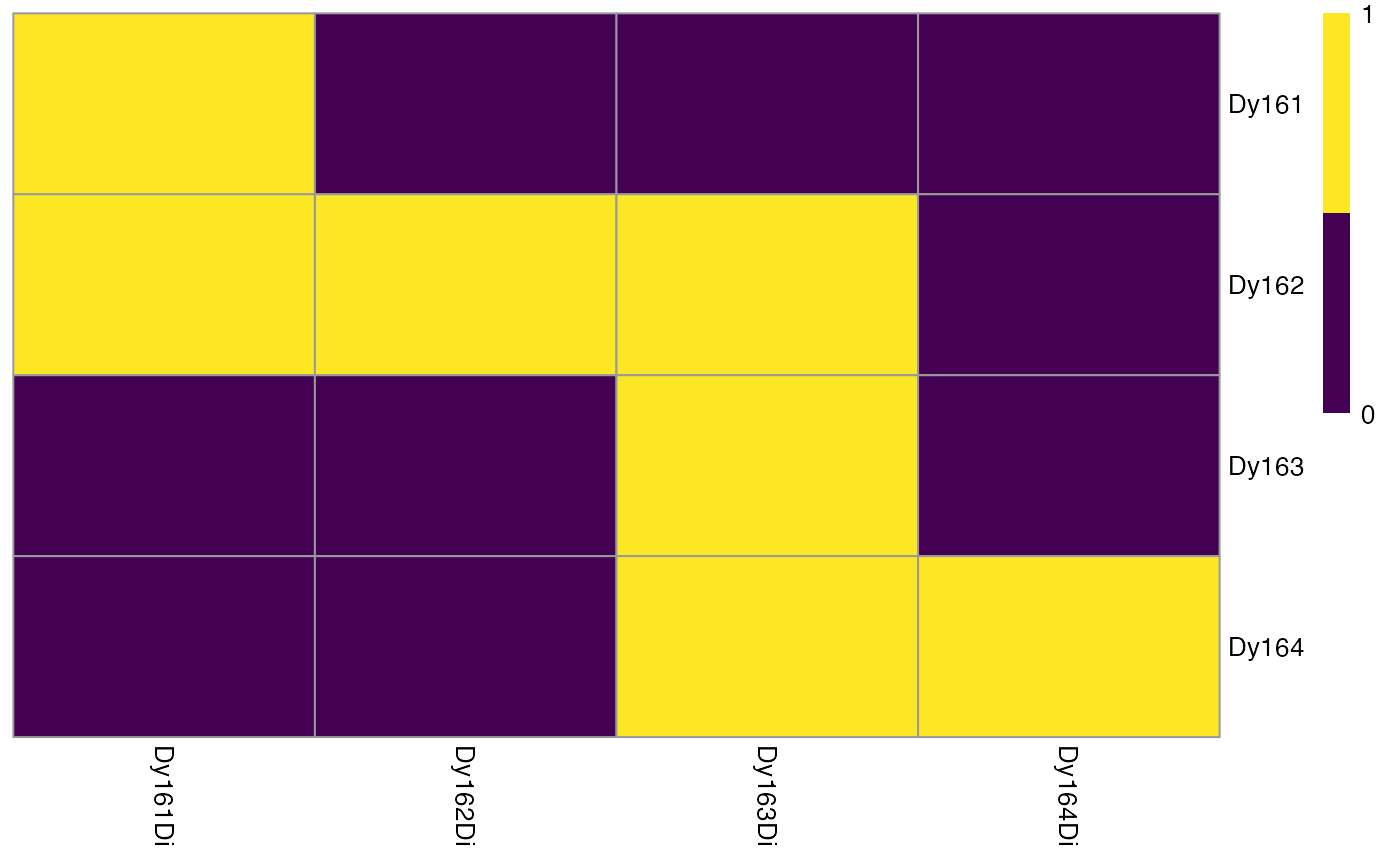
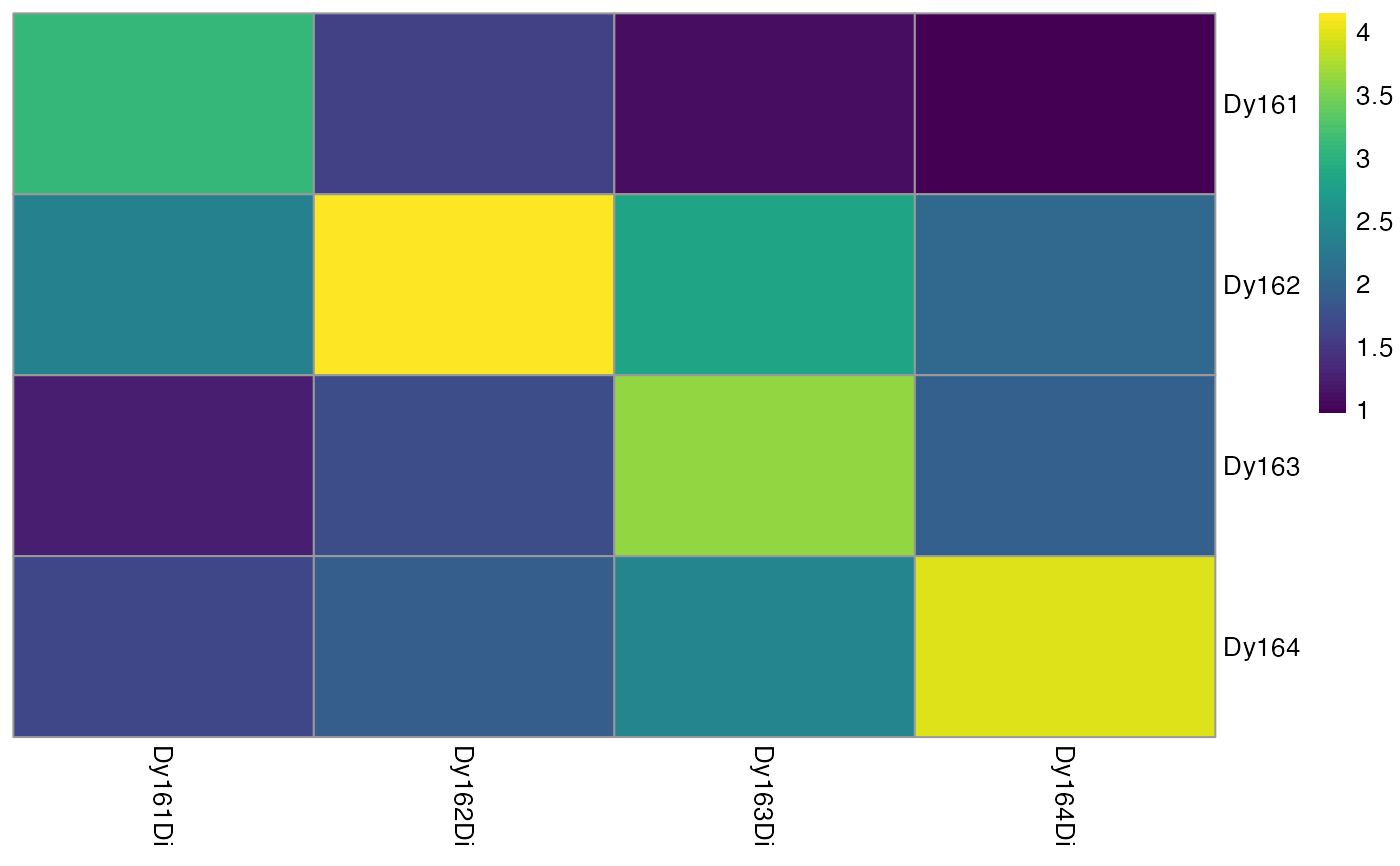
Summarizes and visualizes the pixel intensities per spot and channel
Source:R/plotSpotHeatmap.R
plotSpotHeatmap.RdHelper function for estimating the spillover matrix. This function visualizes the median pixel intensities per spot (rows) and per channel (columns) in form of a heatmap.
plotSpotHeatmap(
object,
spot_id = "sample_id",
channel_id = "channel_name",
assay_type = "counts",
statistic = "median",
log = TRUE,
threshold = NULL,
order_metals = TRUE,
color = viridis(100),
breaks = NA,
legend_breaks = NA,
cluster_cols = FALSE,
cluster_rows = FALSE,
...
)Arguments
- object
a
SingleCellExperimentobject containing pixel intensities per channel. Individual pixels are stored as columns and channels are stored as rows.- spot_id
character string indicating which
colData(object)entry stores the isotope names of the spotted metal. Entries should be of the form (mt)(mass) (e.g. Sm152 for Samarium isotope with the atomic mass 152).- channel_id
character string indicating which
rowData(object)entry contains the isotope names of the acquired channels.- assay_type
character string indicating which assay to use (default
counts).- statistic
the statistic to use when aggregating channels per spot (default
median)- log
should the aggregated pixel intensities be
log10(x + 1)transformed?- threshold
single numeric indicating a threshold after pixel aggregation. All aggregated values larger than
thresholdwill be labeled as1.- order_metals
should the metals be ordered based on spotted mass?
- color
see parameter in
pheatmap- breaks
see parameter in
pheatmap- legend_breaks
see parameter in
pheatmap- cluster_cols
see parameter in
pheatmap- cluster_rows
see parameter in
pheatmap- ...
other arguments passed to
pheatmap.
Value
a pheatmap object
Quality control for spillover estimation
Visualizing the aggregated pixel intensities serves two purposes:
Small median pixel intensities (< 200 counts) might hinder the robust estimation of the channel spillover. In that case, consecutive pixels can be summed (see
binAcrossPixels).Each spotted metal (row) should show the highest median pixel intensity in its corresponding channel (column). If this is not the case, either the naming of the .txt files was incorrect or the incorrect metal was spotted.
By setting the threshold parameter, the user can easily identify spots
where pixel intensities are too low for robust spillover estimation.
See also
pheatmap for visual modifications
aggregateAcrossCells for the aggregation
function
Examples
path <- system.file("extdata/spillover", package = "imcRtools")
# Read in .txt files
sce <- readSCEfromTXT(path)
#> Spotted channels: Dy161, Dy162, Dy163, Dy164
#> Acquired channels: Dy161, Dy162, Dy163, Dy164
#> Channels spotted but not acquired:
#> Channels acquired but not spotted:
# Visualizes heatmap
plotSpotHeatmap(sce)
 # Visualizes thresholding results
plotSpotHeatmap(sce, log = FALSE, threshold = 200)
# Visualizes thresholding results
plotSpotHeatmap(sce, log = FALSE, threshold = 200)