Function to visualize cell-level information on segmentation masks
Source:R/plotCells.R
plotCells.RdThis function takes a SingleCellExperiment and
CytoImageList object containing segmentation masks to
colour cells by marker expression or metadata.
plotCells(
mask,
object = NULL,
cell_id = NULL,
img_id = NULL,
colour_by = NULL,
outline_by = NULL,
exprs_values = "counts",
colour = NULL,
...
)Arguments
- mask
a
CytoImageListcontaining single-channelImageobjects (see section 'Segmentation mask object' below).- object
a
SingleCellExperimentobject.- cell_id
character specifying the
colData(object)entry, in which the integer cell IDs are stored. These IDs should match the integer pixel values in the segmentation mask object.- img_id
character specifying the
colData(object)andmcols(mask)entry, in which the image IDs are stored (see section 'Linking theSingleCellExperimentandCytoImageListobject' below)- colour_by
character or character vector specifying the features (
rownames(object)) or metadata (colData(object)entry) used to colour individual cells. Cells can be coloured by singlecolData(object)entries or by up to six features.- outline_by
single character indicating the
colData(object)entry by which to outline individual cells.- exprs_values
single character indicating which
assay(object)entry to use when visualizing feature counts.- colour
a list with names matching the entries to
colour_byand/oroutline_by. When setting the colour for continous features, at least two colours need to be provided indicating the colours for minimum and maximum values. When colouring discrete vectors, a colour for each unique entry needs to be provided (see section 'Setting the colours' and examples)- ...
Further arguments passed to
?"plotting-param"
Value
a list if return_images and/or return_plot is TRUE
(see ?"plotting-param").
plot: a single plot object (display = "all") or a list of plot objects (display = "single")images: aSimpleListobject containing three-colourImageobjects.
Segmentation mask object
In the plotCells function, mask refers to a
CytoImageList object that contains a single or multiple
segmentation masks in form of individual Image objects.
The function assumes that each object in the segmentation mask is a cell.
The key features of such masks include:
each Image object contains only one channel
pixel values are integers indicating the cells' IDs or 0 (background)
Linking SingleCellExperiment and CytoImageList objects
To colour individual cells contained in the segmentation masks based on
features and metadata stored in the SingleCellExperiment object, an
img_id and cell_id entry needs to be provided. Image IDs are
matched between the SingleCellExperiment and CytoImageList
object via entries to the colData(object)[,img_id] and the
mcols(mask)[,img_id] slots. Cell IDs are matched between the
SingleCellExperiment and CytoImageList object via entries to
colData(object)[,cell_id] and the integer values of the segmentation
masks.
Setting the colours
By default, features and metadata are coloured based on internally-set
colours. To set new colours, a list object must be provided. The names
of the object must correspond to the entries to colour_by and/or
outline_by. When setting the colours for continous expression values
or continous metadata entries, a vector of at least two colours need to be
specified. These colours will be passed onto colorRampPalette
for interpolation. Discrete metadata entries can be coloured by specifying a
named vector in which each entry corresponds to a unique entry to the
metadata vector.
Subsetting the CytoImageList object
The CytoImageList object can be subsetted before calling the
plotCells function. In that case, only the selected images are
displayed.
Subsetting the SingleCellExperiment object
The SingleCellExperiment object can be subsetted before calling the
plotCells function. In that case, only cells contained in the
SingleCellExperiment object are coloured/outlined.
Colour scaling
When colouring features using the plotCells function, colours are scaled between the minimum and maximum per feature across the full assay contained in the SingleCellExperiment object. When subsetting images, cell-level expression is not scaled across the subsetted images but the whole SingleCellExperiment object. To avoid this, the SingleCellExperiment object can be subsetted to only contain the cells that should be displayed before plotting.
See also
For further plotting parameters see ?"plotting-param"
Examples
data(pancreasMasks)
data(pancreasSCE)
# Visualize the masks
plotCells(pancreasMasks)
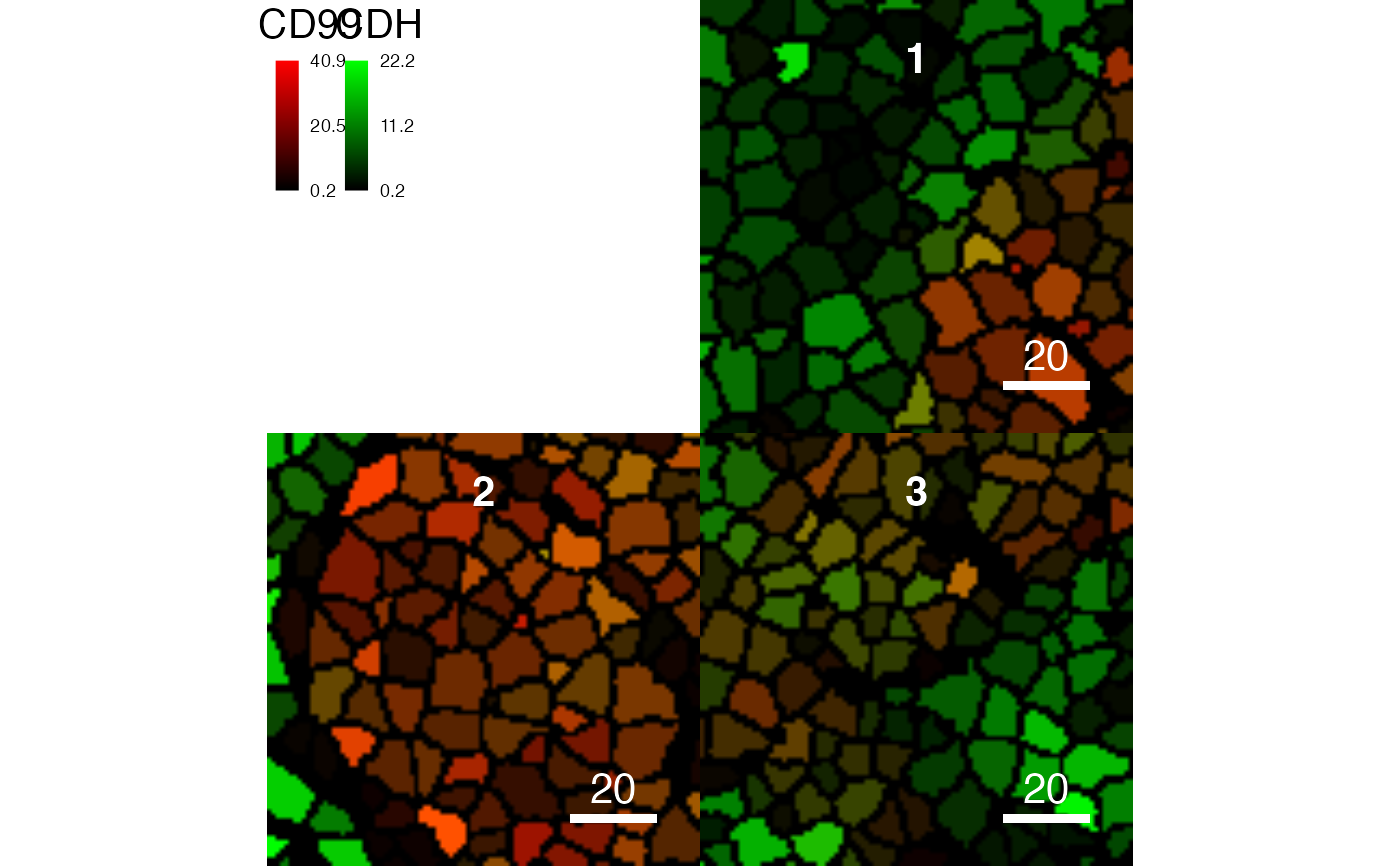
 # Colour the masks based on averaged expression
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = c("CD99", "CDH"))
# Colour the masks based on averaged expression
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = c("CD99", "CDH"))
 # Colour the masks based on metadata
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CellType")
# Colour the masks based on metadata
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CellType")
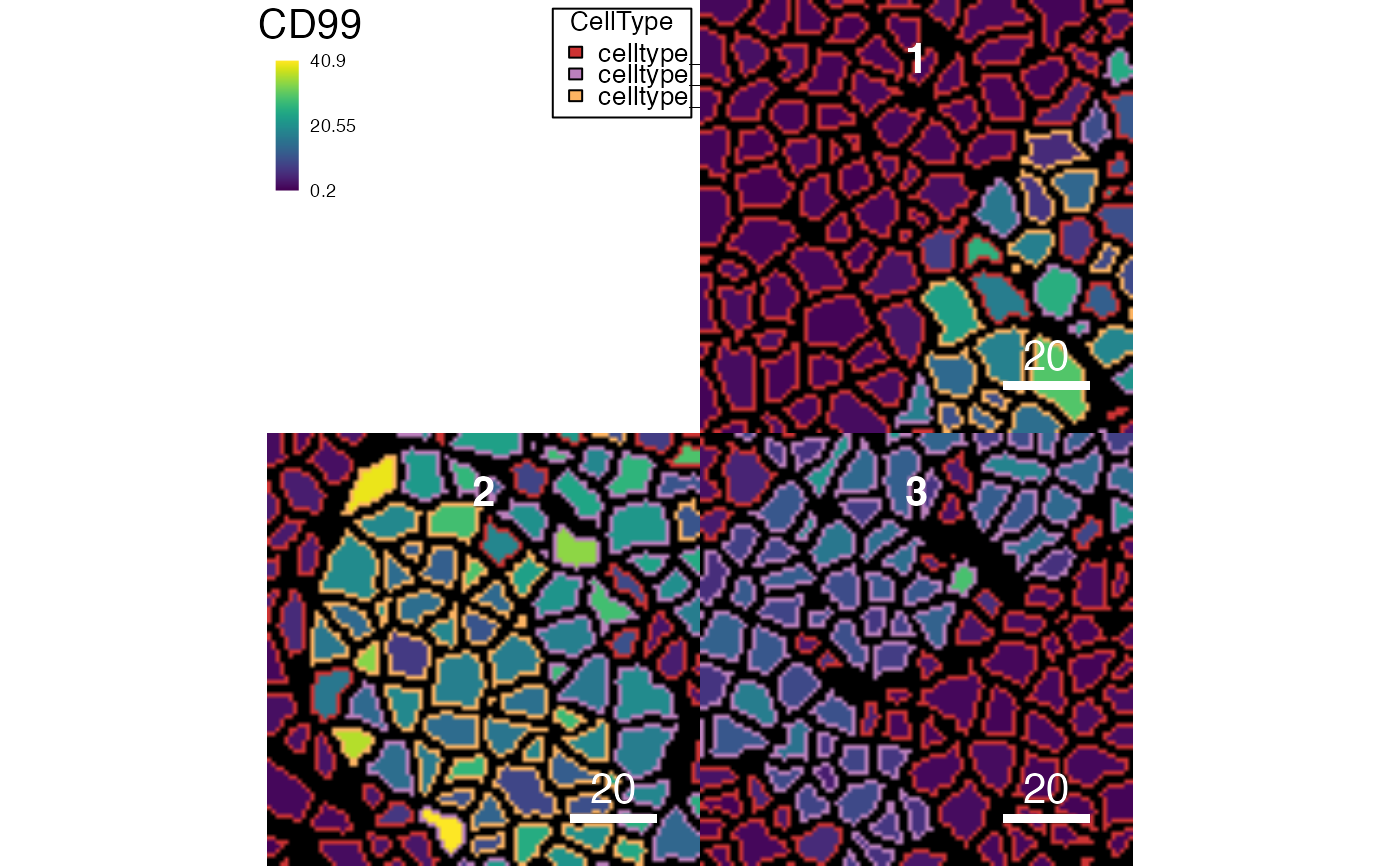
 # Outline the masks based on metadata
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99",
outline_by = "CellType")
# Outline the masks based on metadata
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99",
outline_by = "CellType")
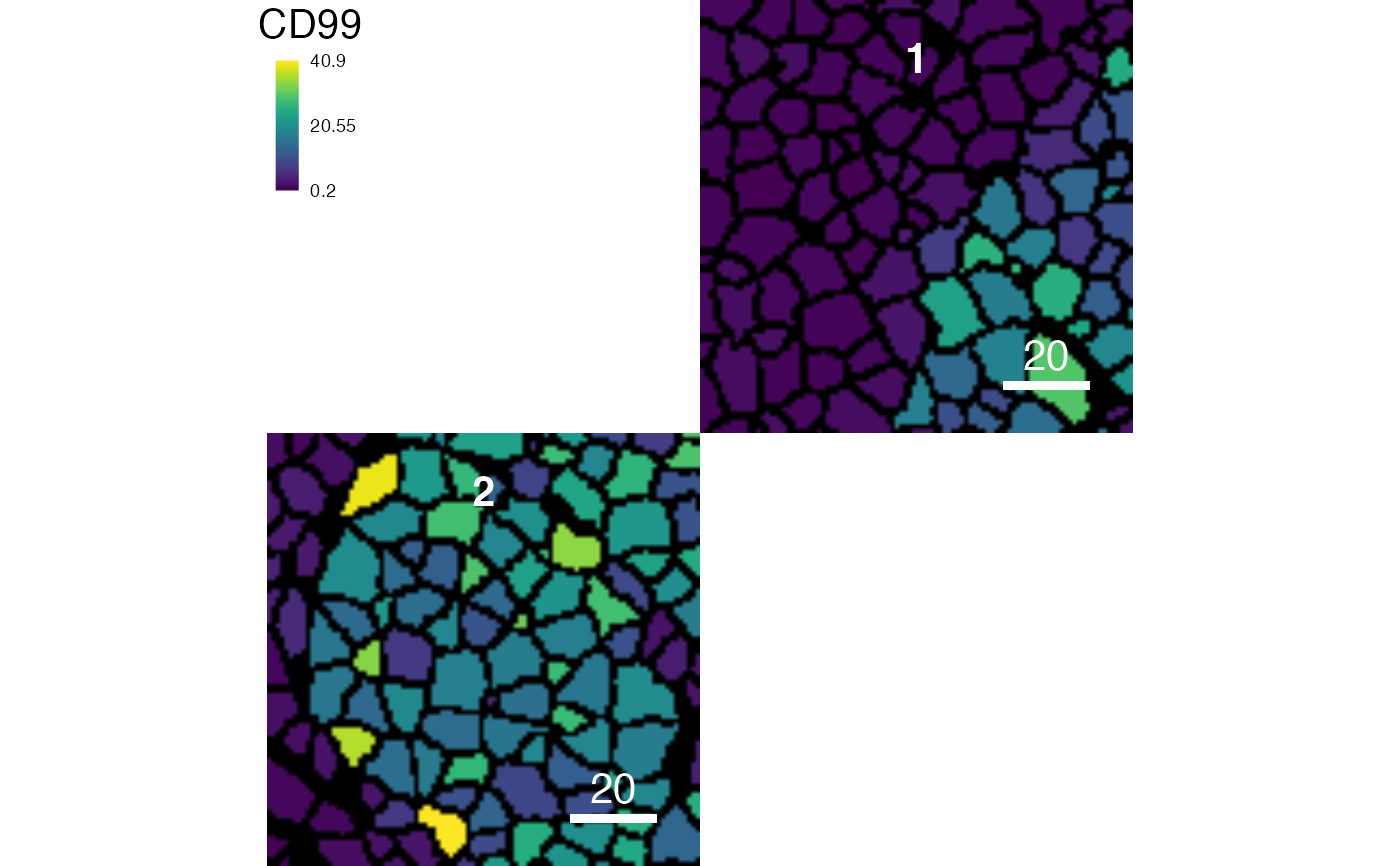
 # Colour the masks based on arcsinh-transformed expression
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99",
exprs_values = "exprs")
# Colour the masks based on arcsinh-transformed expression
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99",
exprs_values = "exprs")
 # Subset the images
cur_images <- getImages(pancreasMasks, 1:2)
plotCells(cur_images, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99")
# Subset the images
cur_images <- getImages(pancreasMasks, 1:2)
plotCells(cur_images, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99")
 # Set colour
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99", outline_by = "CellType",
colour = list(CD99 = c("black", "red"),
CellType = c(celltype_A = "blue",
celltype_B = "green",
celltype_C = "red")))
# Set colour
plotCells(pancreasMasks, object = pancreasSCE, img_id = "ImageNb",
cell_id = "CellNb", colour_by = "CD99", outline_by = "CellType",
colour = list(CD99 = c("black", "red"),
CellType = c(celltype_A = "blue",
celltype_B = "green",
celltype_C = "red")))
