The plotCells and plotPixels functions share a
number of parameter that can be set to change the visual representation of
plotted images.
Arguments
- missing_colour
a single character specifying a valid colour. Cells that are not contained in the SingleCellExperiment object will be coloured based on
missing_colour. In theplotPixelsfunction,missing_colourdefines the outline of the cells ifoutline_byis not set.- background_colour
(only
plotCells) a single character specifying a valid colour that is set as the background of the image.- scale_bar
a list specifying features of the scale bar. One or multiple of the following entries are supported:
length: numeric length in pixels (default 20% of the largest image width).label: single character specifying the scale bar label.cex: numeric value indicating the size of the scale bar label.lwidth: numeric value indicating the line width of the scale bar in pixels. By default, the line width is adjusted relative to the maximum height of the images.colour: single character specifying the colour of scale bar and label (default "white").position: position of scale bar. Supported values: "topleft", "topright", "bottomleft", "bottomright" (default "bottomright").margin: vector of two numeric entries specifying the x and y margin between image boundary and the scale bar (default c(10,10)).frame: either "all" to display scale bar on all images or a single number specifying the image for which the scale bar should be displayed (default "all").
Plotting of the scale bar is suppressed if set to
NULL.- image_title
a list specifying features of the image titles. One or multiple of the following entries are supported:
text: character vector of image titles. Same length as theCytoImageListobject.position: single character specifying the position of the title. Supported entries: "top", "bottom", "topleft", "bottomleft", "topright", "bottomright" (default "top").colour: single character specifying the colour of image title (default "white").margin: vector of two numeric entries specifying the x and y margin between image boundary and the image title (default c(10,10)).font: numeric entry specifying the font of the image title (default 1, seeparfor details)cex: numeric value indicating the size of the image title.
Plotting of the image title is suppressed if set to
NULL.- save_plot
a list specifying how to save the plot. One or multiple of the following entries are supported:
filename: single character specifying a valid file name. The file extension specifies the format in which the file is saved. Supported formats are: jpeg, tiff and png. Ifdisplay = "single", each image will be written in an individual file. The file names obtain a "_x" ending where x indicates the position of the image in theCytoImageListobject or "legend".scale: by default the height and width of the saved image is defined by the maximum image size times the number of rows and number of columns. This resolution is often not sufficient to clearly display the text. Thescaleparameter can be set to increase the resolution of the image while keeping the text size constant (default 1 but can be increased for optimal results).
- return_plot
logical indicating whether to return the plot (see
recordPlotfor more infos).- return_images
logical indicating whether to return the coloured images in form of a
SimpleListobject. Each entry to this list is a three-colourImageobject. However, the image title and scale bar are not retained.- legend
a list specifying features of the legend. One or multiple of the following entries are supported:
colour_by.title.font: numeric entry specifying the font of the legend title for features specified bycolour_by.colour_by.title.cex: numeric entry specifying the size of the legend title for features specified bycolour_by.colour_by.labels.cex: numeric entry specifying the size of the legend labels for features specified bycolour_by.colour_by.legend.cex: (only discrete features) numeric entry specifying the size of the legend for features specified bycolour_by.outline_by.title.font: numeric entry specifying the font of the legend title for features specified byoutline_by.outline_by.title.cex: numeric entry specifying the size of the legend title for features specified byoutline_by.outline_by.labels.cex: numeric entry specifying the size of the legend labels for features specified byoutline_by.outline_by.legend.cex: (only discrete features) numeric entry specifying the size of the legend for features specified byoutline_by.margin: numeric value indicating the margin (in pixels) between the legends and the outer boundary (default 2)
Plotting of the legend is suppressed if set to
NULL.- margin
numeric value indicating the gap (in pixels) between individual images (default 0).
- thick
logical indicating whether cell borders should be drawn as thick lines (default
FALSE).- display
one of two possible values: "all" or "single". When set to "all", all images are displayed at once in a grid-like fashion. When set to "single", individual images are plotted in single graphics devices. The second option is useful when saving individual images in pdf format or when displaying in Rmarkdown files.
- scale
logical indicating whether to scale each feature individually to its minimum/maximum across the SingleCellExperiment object (see
plotCells) or across all displayed images (seeplotCells). If set toFALSEeach value is displayed relative to the maximum of all selected features.- interpolate
a logical indicating whether to apply linear interpolation to the image when drawing (see
rasterImage) (default TRUE).
Value
a list if return_images and/or return_plot is TRUE.
plot: a single plot object (display = "all") or a list of plot objects (display = "single")images: aSimpleListobject containing three-colourImageobjects.
See also
plotCells and plotPixels
for the main plotting functions
Examples
data("pancreasImages")
data("pancreasMasks")
data("pancreasSCE")
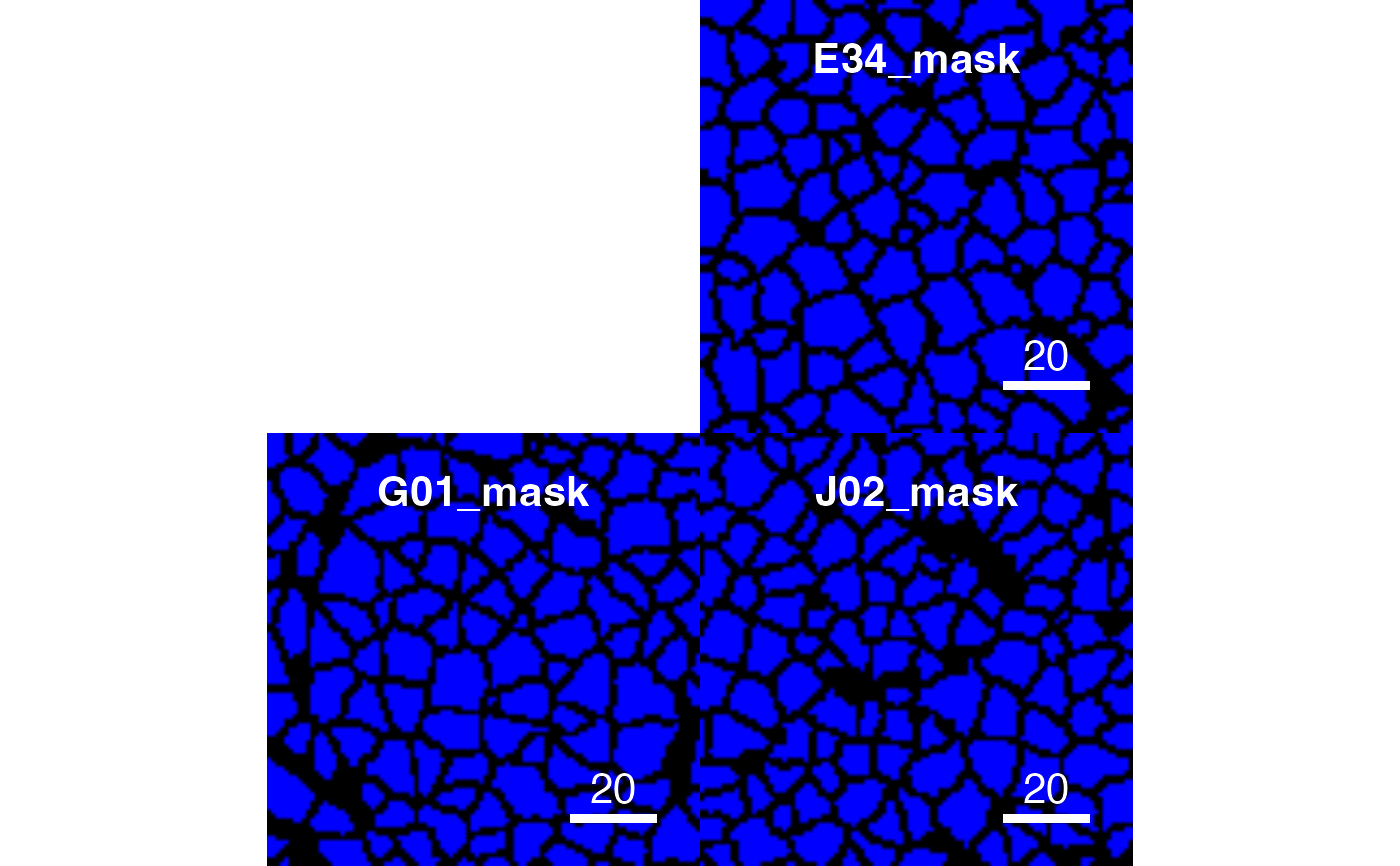
# Setting missing colour
plotCells(pancreasMasks, missing_colour = "blue")
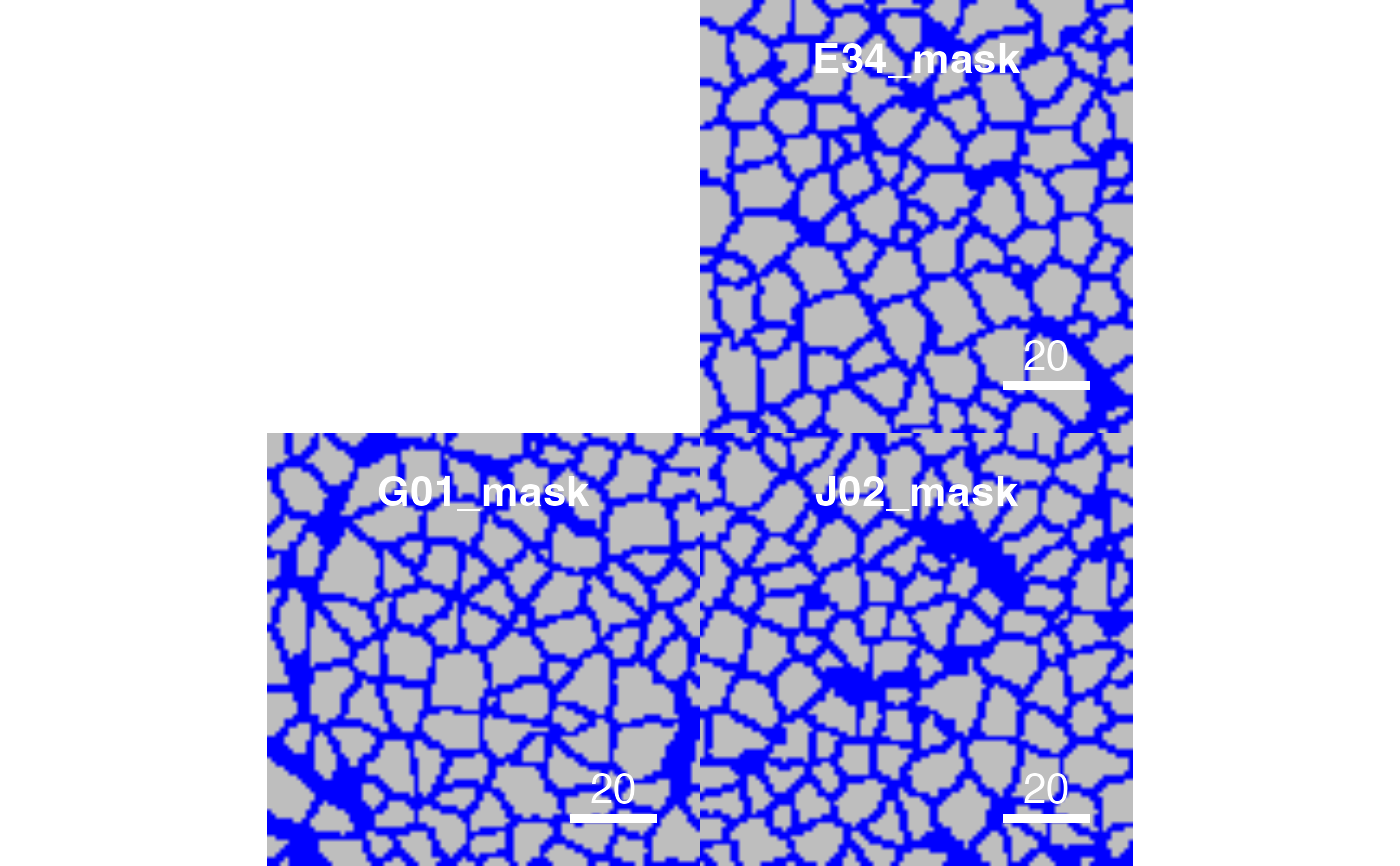
 # Setting background colour
plotCells(pancreasMasks, background_colour = "blue")
# Setting background colour
plotCells(pancreasMasks, background_colour = "blue")
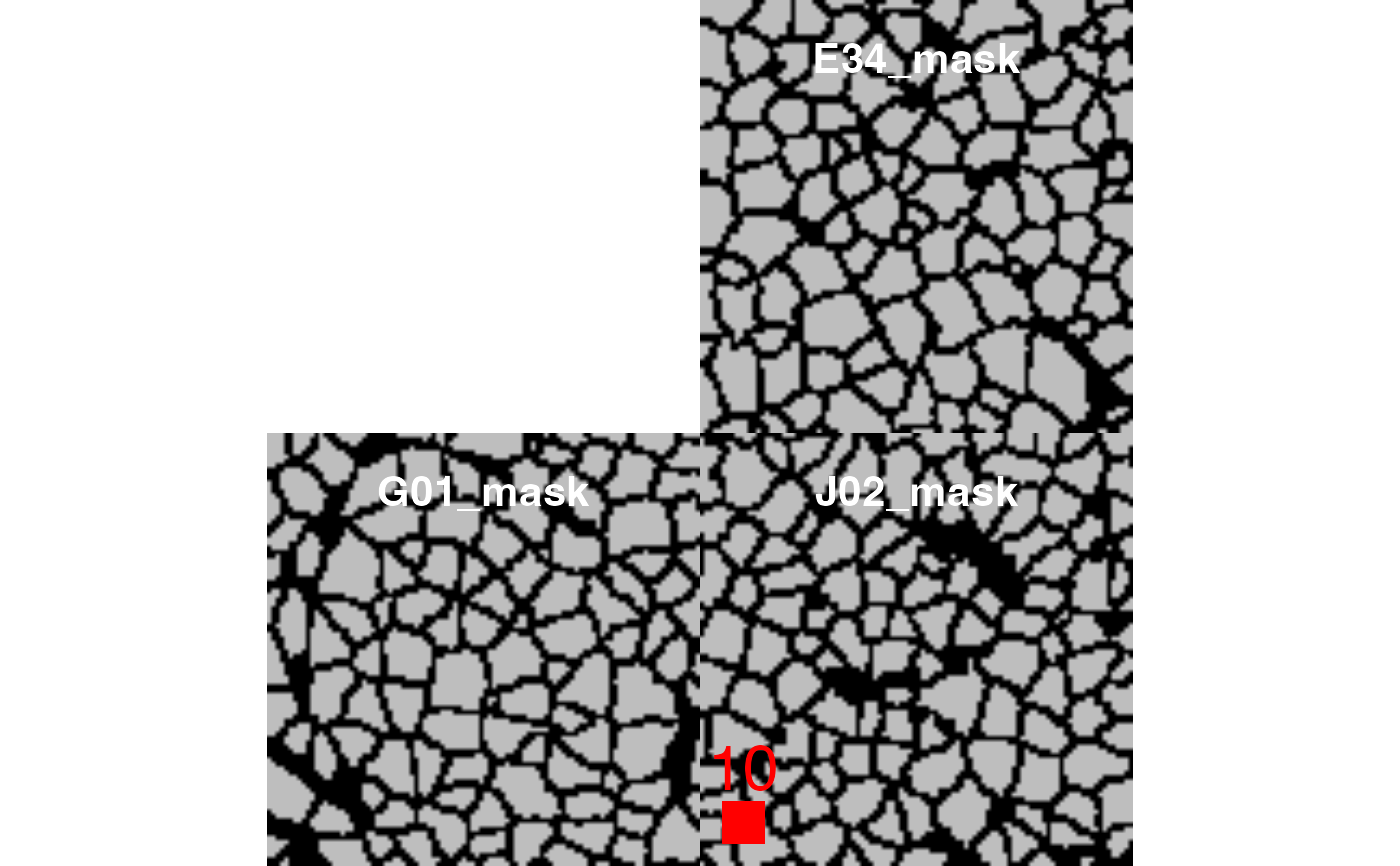
 # Setting the scale bar
plotCells(pancreasMasks, scale_bar = list(length = 10,
cex = 2,
lwidth = 10,
colour = "red",
position = "bottomleft",
margin = c(5,5),
frame = 3))
# Setting the scale bar
plotCells(pancreasMasks, scale_bar = list(length = 10,
cex = 2,
lwidth = 10,
colour = "red",
position = "bottomleft",
margin = c(5,5),
frame = 3))
 # Setting the image title
plotCells(pancreasMasks,
image_title = list(text = c("image1", "image2", "image3"),
position = "topleft",
colour = "blue",
margin = c(0,5),
font = 2,
cex = 2))
# Setting the image title
plotCells(pancreasMasks,
image_title = list(text = c("image1", "image2", "image3"),
position = "topleft",
colour = "blue",
margin = c(0,5),
font = 2,
cex = 2))
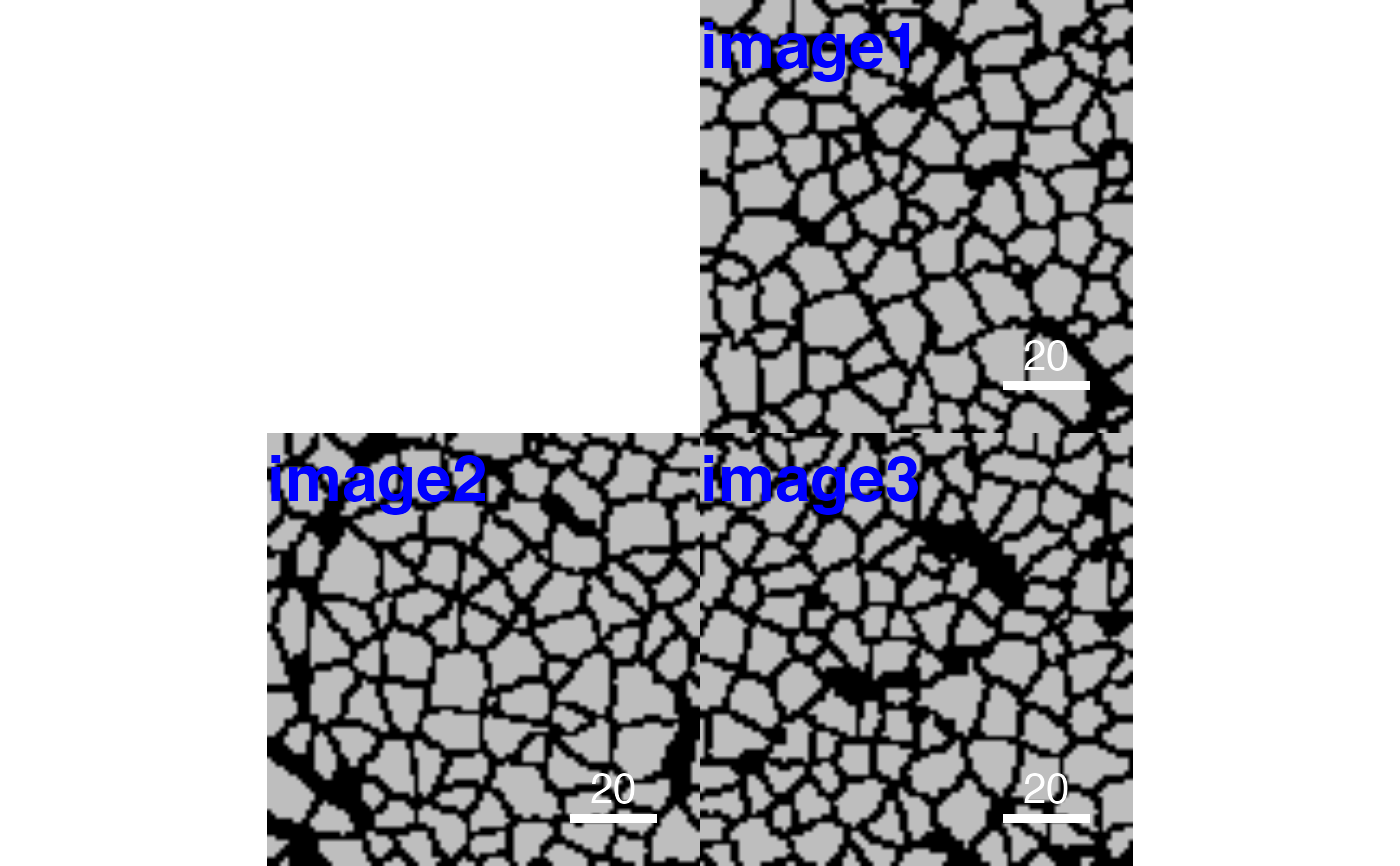 # Return plot
cur_out <- plotPixels(pancreasImages, return_plot = TRUE)
# Return plot
cur_out <- plotPixels(pancreasImages, return_plot = TRUE)
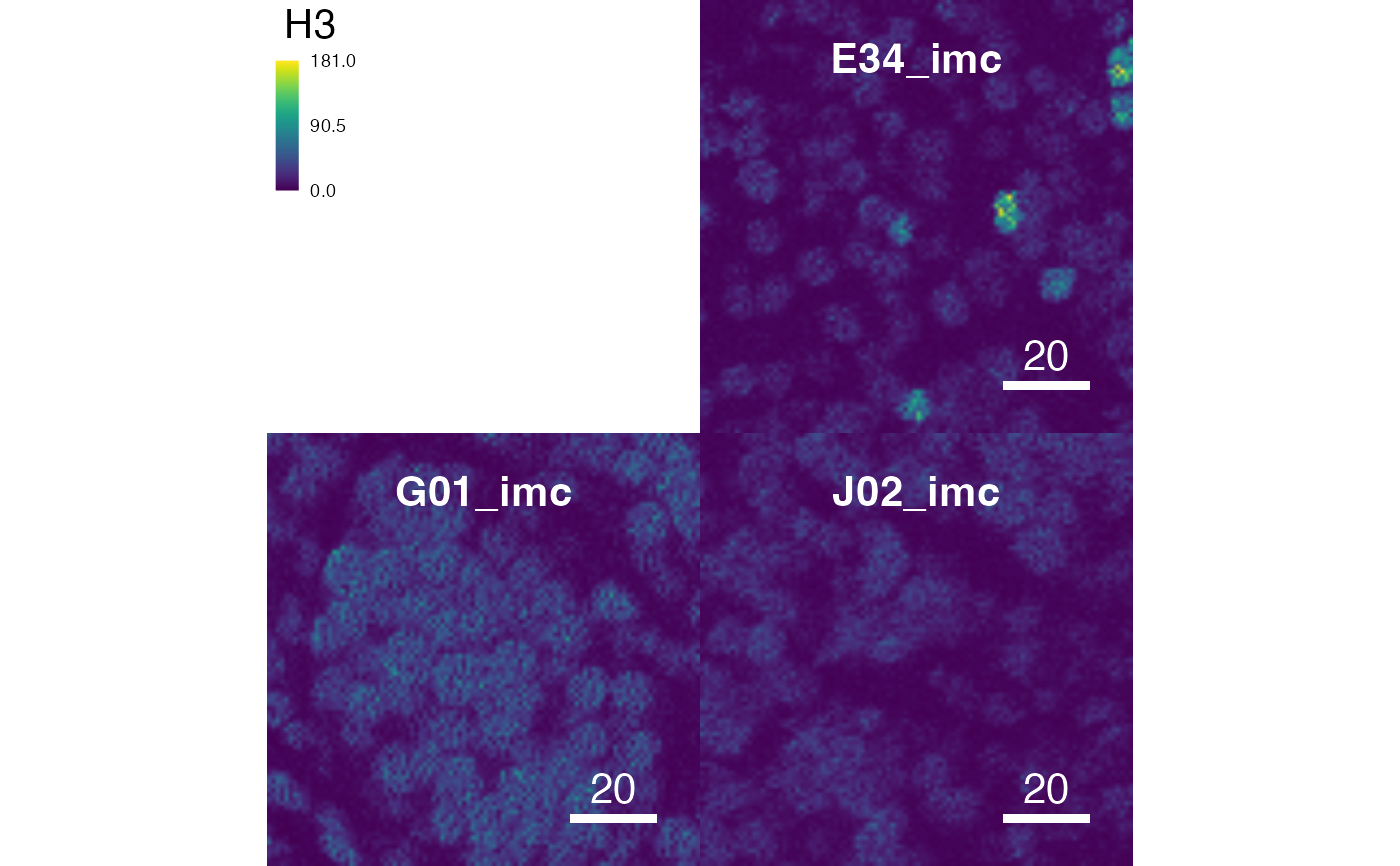 cur_out$plot
cur_out$plot
 # Return images
cur_out <- plotPixels(pancreasImages, return_images = TRUE)
cur_out$images
#> List of length 3
#> names(3): E34_imc G01_imc J02_imc
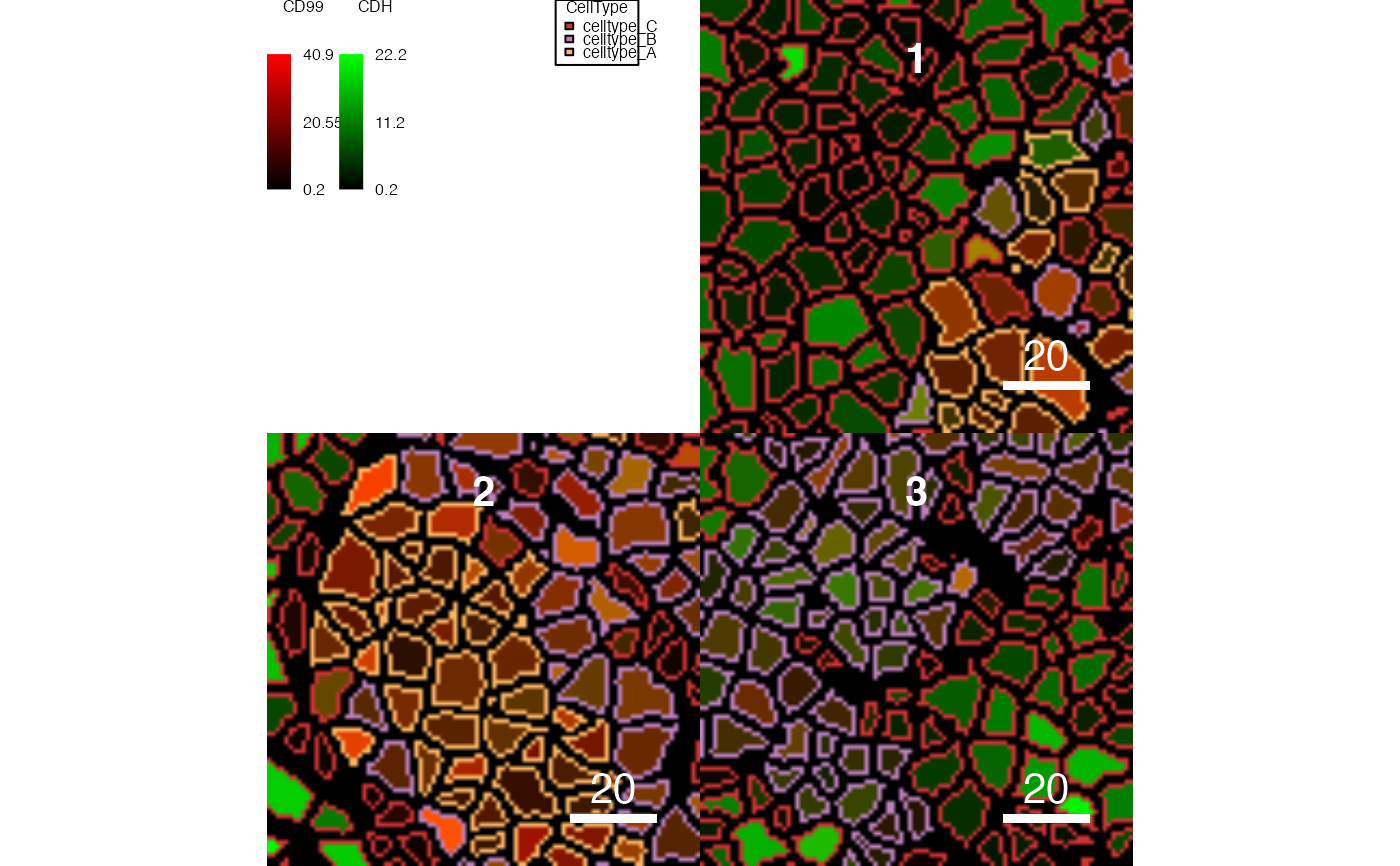
# Setting the legend
plotCells(pancreasMasks, object = pancreasSCE,
img_id = "ImageNb", cell_id = "CellNb",
colour_by = c("CD99", "CDH"),
outline_by = "CellType",
legend = list(colour_by.title.font = 0.5,
colour_by.title.cex = 0.5,
colour_by.labels.cex = 0.5,
outline_by.legend.cex = 0.5,
margin = 0))
# Return images
cur_out <- plotPixels(pancreasImages, return_images = TRUE)
cur_out$images
#> List of length 3
#> names(3): E34_imc G01_imc J02_imc
# Setting the legend
plotCells(pancreasMasks, object = pancreasSCE,
img_id = "ImageNb", cell_id = "CellNb",
colour_by = c("CD99", "CDH"),
outline_by = "CellType",
legend = list(colour_by.title.font = 0.5,
colour_by.title.cex = 0.5,
colour_by.labels.cex = 0.5,
outline_by.legend.cex = 0.5,
margin = 0))
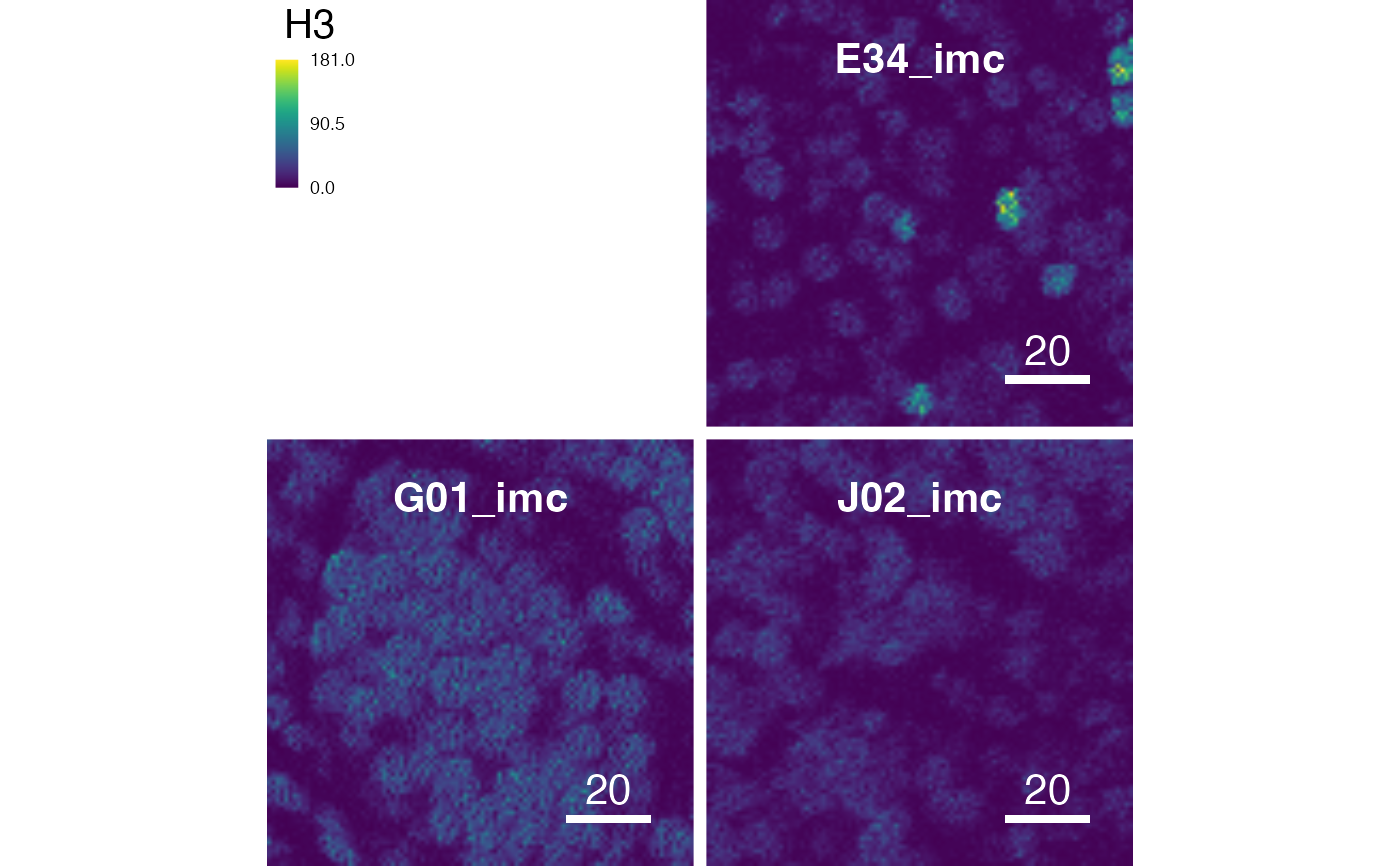
 # Setting the margin between images
plotPixels(pancreasImages, margin = 3)
# Setting the margin between images
plotPixels(pancreasImages, margin = 3)
 # Thick outlines
plotCells(pancreasMasks, object = pancreasSCE,
img_id = "ImageNb", cell_id = "CellNb",
colour_by = "CD99",
outline_by = "CellType",
thick = TRUE)
# Thick outlines
plotCells(pancreasMasks, object = pancreasSCE,
img_id = "ImageNb", cell_id = "CellNb",
colour_by = "CD99",
outline_by = "CellType",
thick = TRUE)
 # Displaying individual images
plotPixels(pancreasImages, display = "single")
# Displaying individual images
plotPixels(pancreasImages, display = "single")
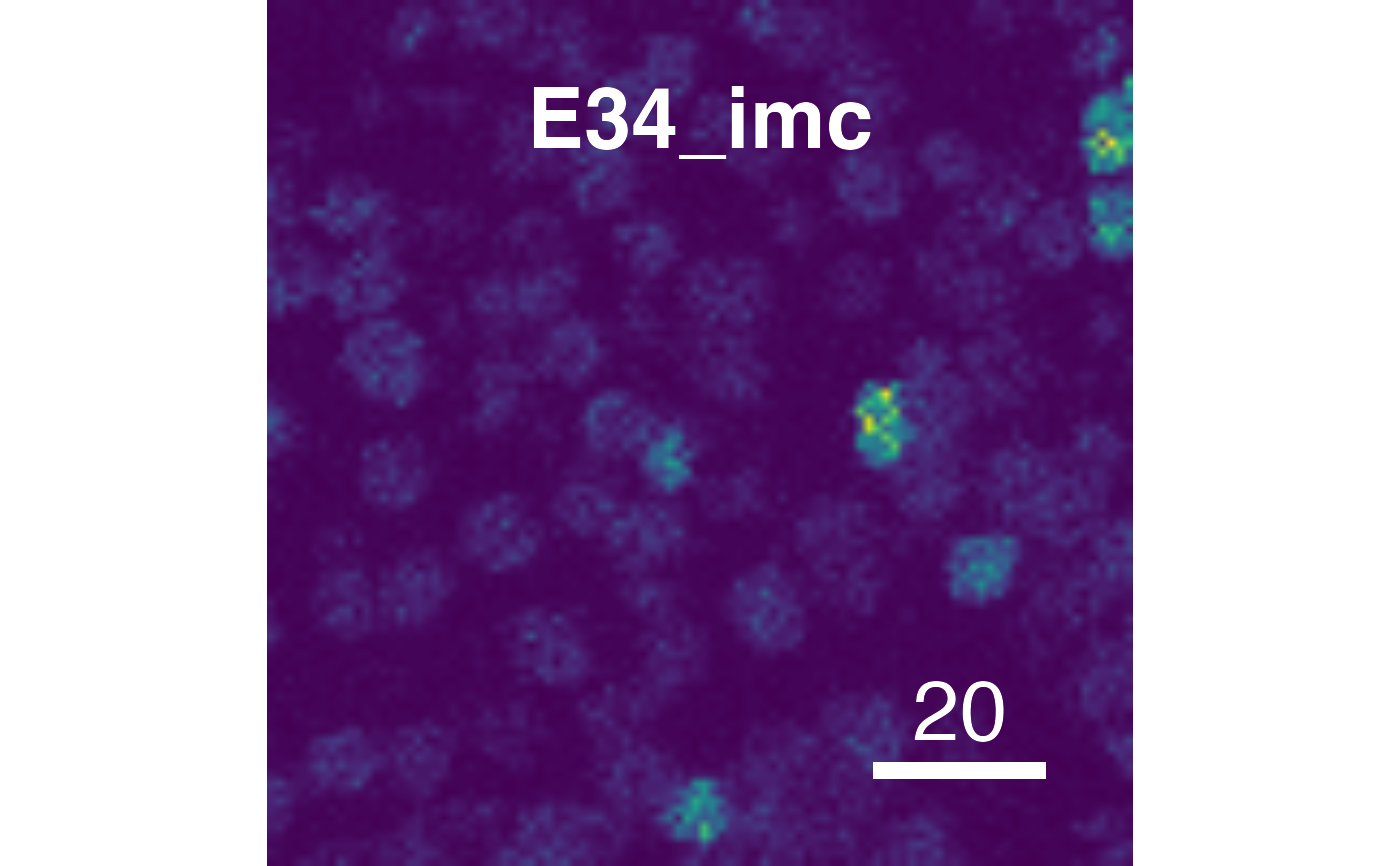
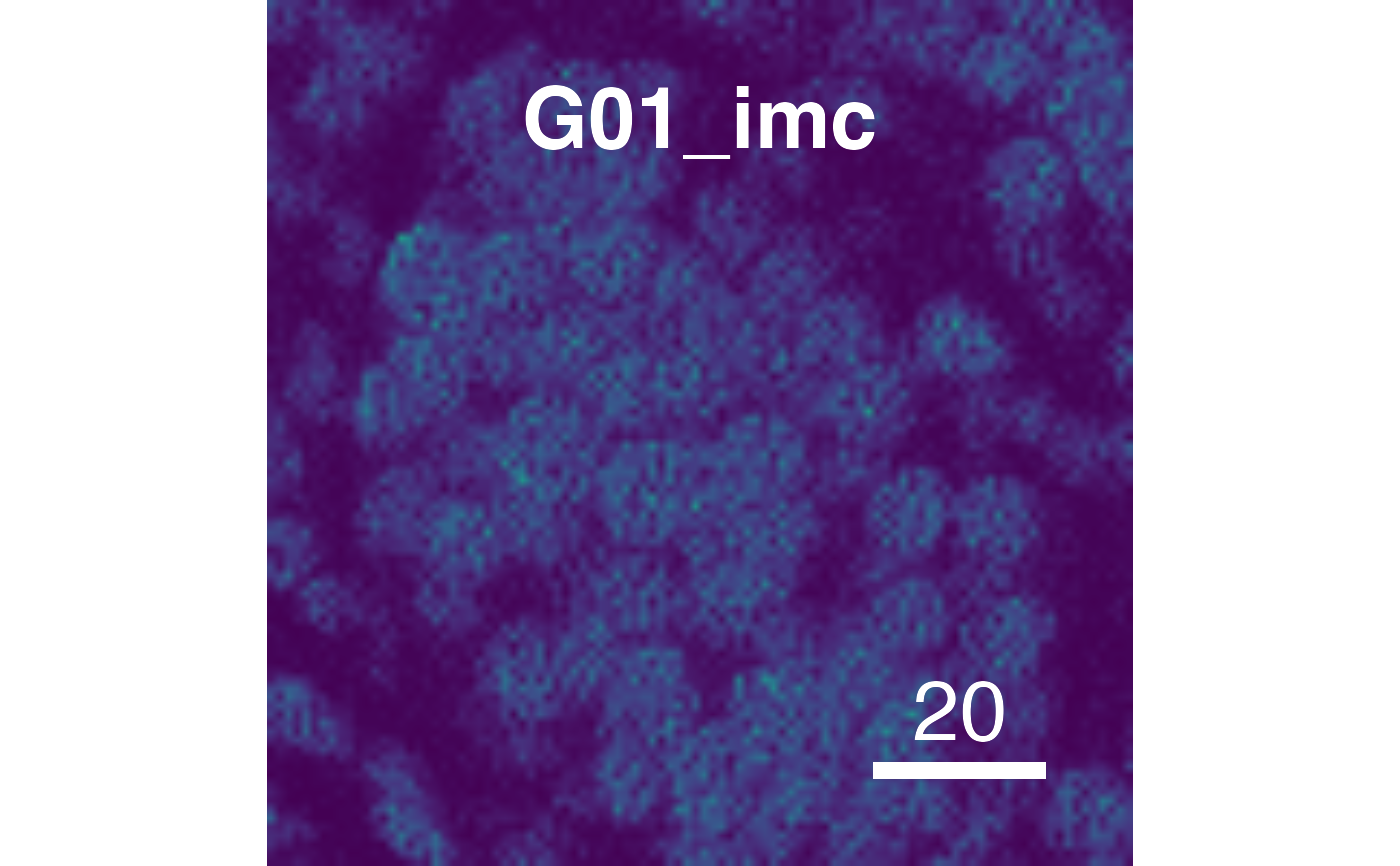
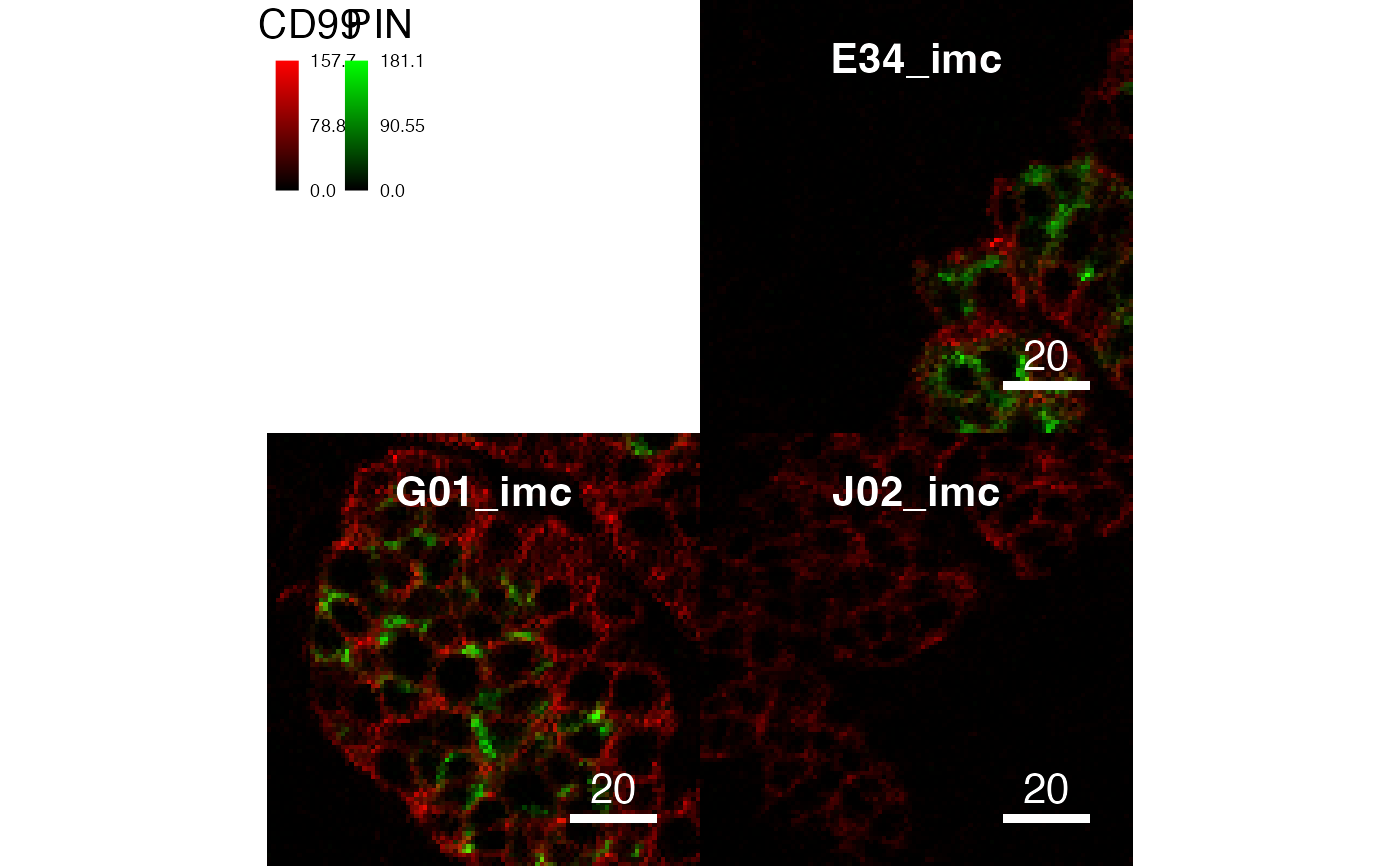
 # Supress scaling
plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
scale = TRUE)
# Supress scaling
plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
scale = TRUE)
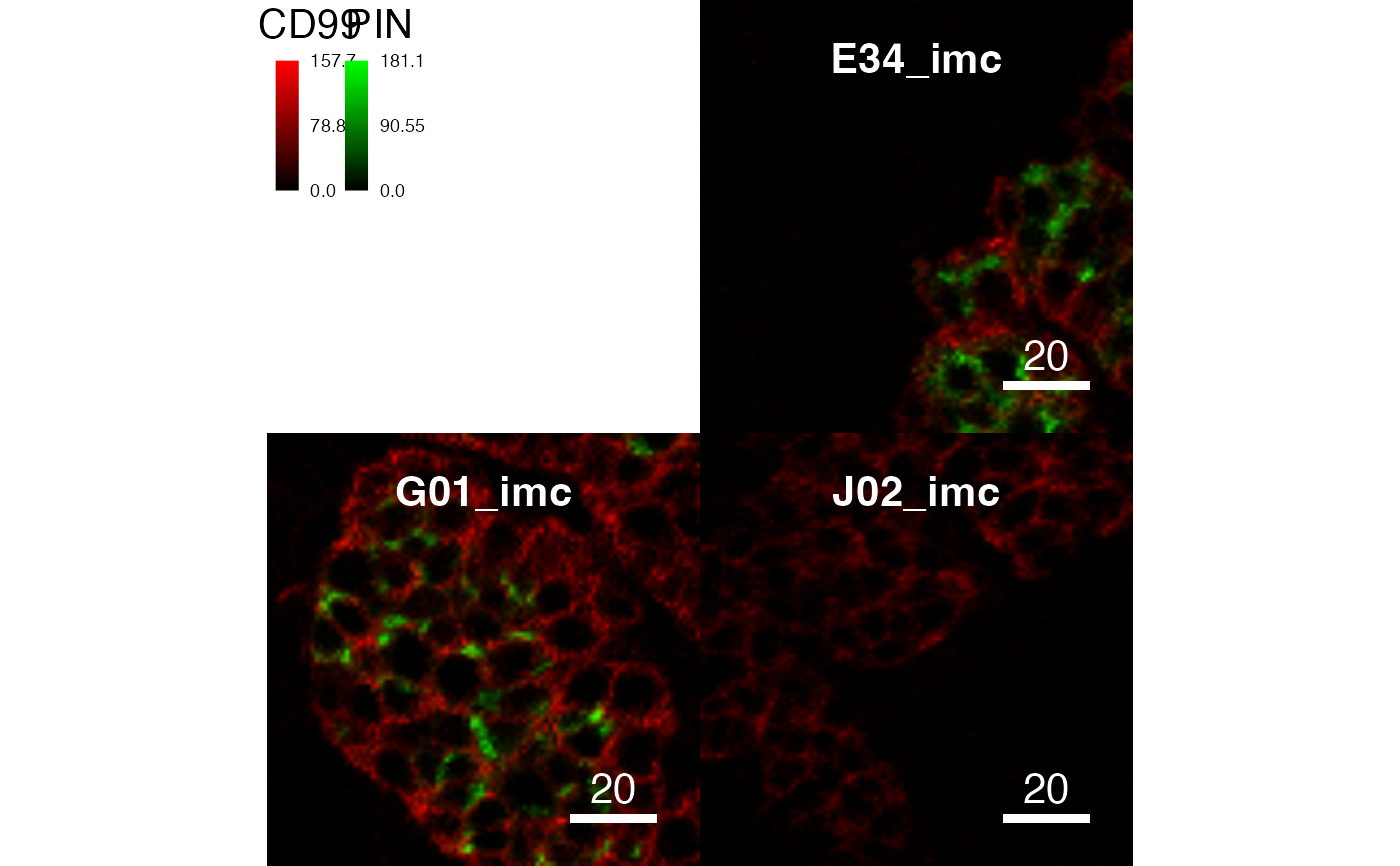 plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
scale = FALSE)
plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
scale = FALSE)
 # Suppress interpolation
plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
interpolate = TRUE)
# Suppress interpolation
plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
interpolate = TRUE)
 plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
interpolate = FALSE)
plotPixels(pancreasImages, colour_by = c("CD99", "PIN"),
interpolate = FALSE)