Function to plot directed spatial context graphs based on symbolic edge-lists and vertex metadata, which operates on the cohort-level. The user can specify node, node_label and edge aesthetics.
plotSpatialContext(
object,
entry = "spatial_context",
group_by = "sample_id",
node_color_by = NULL,
node_size_by = NULL,
node_color_fix = NULL,
node_size_fix = NULL,
node_label_repel = TRUE,
node_label_color_by = NULL,
node_label_color_fix = NULL,
draw_edges = TRUE,
edge_color_fix = NULL,
return_data = FALSE
)Arguments
- object
a
SingleCellExperimentorSpatialExperimentobject.- entry
single character specifying the
colData(object)entry containing thedetectSpatialContextoutput. Defaults to "spatial_context".- group_by
a single character indicating the
colData(object)entry by which SCs are grouped. This is usually the image or patient ID. Defaults to "sample_id".- node_color_by
single character either
NULL, "name","n_cells", "n_group"by which the nodes should be colored.- node_size_by
single character either
NULL, "n_cells","n_group"by which the size of the nodes are defined.- node_color_fix
single character specifying the color of all nodes.
- node_size_fix
single numeric specifying the size of all nodes.
- node_label_repel
should nodes be labelled? Defaults to TRUE.
- node_label_color_by
single character either
NULL, "name","n_cells","n_group"by which the node labels should be colored.- node_label_color_fix
single character specifying the color of all node labels.
- draw_edges
should edges be drawn between nodes? Defaults to TRUE.
- edge_color_fix
single character specifying the color of all edges.
- return_data
should the edge list and vertex metadata for graph construction be returned as a
listof twodata.frames?
Value
returns a ggplot object or a list of two
data.frames.
See also
detectSpatialContext for the function to detect
spatial contexts
filterSpatialContext for the function to filter
spatial contexts
Examples
set.seed(22)
library(cytomapper)
data(pancreasSCE)
## 1. Cellular neighborhood (CN)
sce <- buildSpatialGraph(pancreasSCE, img_id = "ImageNb",
type = "knn",
name = "knn_cn_graph",
k = 5)
#> The returned object is ordered by the 'ImageNb' entry.
sce <- aggregateNeighbors(sce, colPairName = "knn_cn_graph",
aggregate_by = "metadata",
count_by = "CellType",
name = "aggregatedCellTypes")
cur_cluster <- kmeans(sce$aggregatedCellTypes, centers = 3)
sce$cellular_neighborhood <- factor(cur_cluster$cluster)
plotSpatial(sce, img_id = "ImageNb",
colPairName = "knn_cn_graph",
node_color_by = "cellular_neighborhood",
scales = "free")
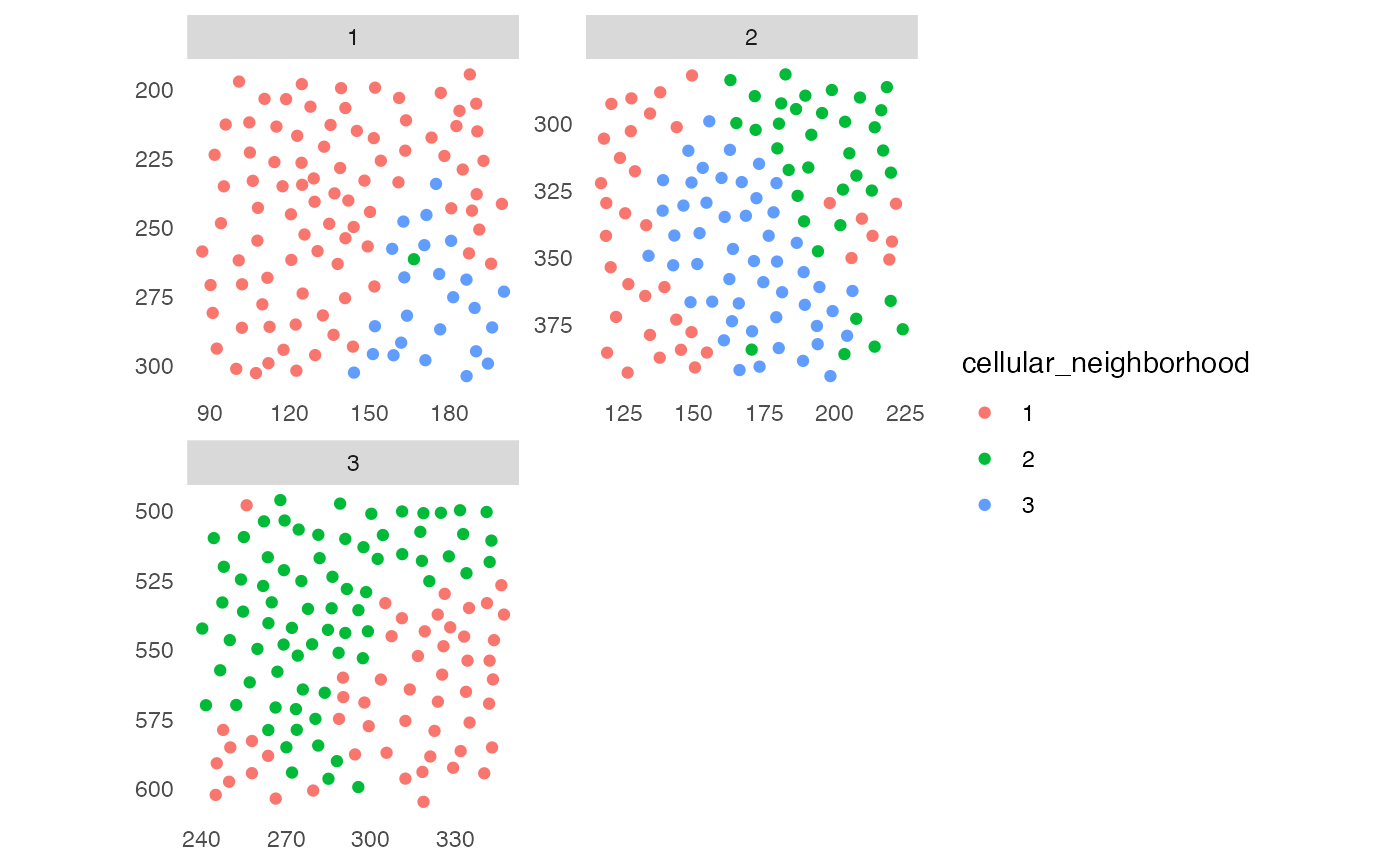 ## 2. Spatial context (SC)
sce <- buildSpatialGraph(sce, img_id = "ImageNb",
type = "knn",
name = "knn_sc_graph",
k = 15)
#> The returned object is ordered by the 'ImageNb' entry.
sce <- aggregateNeighbors(sce, colPairName = "knn_sc_graph",
aggregate_by = "metadata",
count_by = "cellular_neighborhood",
name = "aggregatedNeighborhood")
# Detect spatial context
sce <- detectSpatialContext(sce, entry = "aggregatedNeighborhood",
threshold = 0.9)
plotSpatial(sce, img_id = "ImageNb",
colPairName = "knn_sc_graph",
node_color_by = "spatial_context",
scales = "free")
## 2. Spatial context (SC)
sce <- buildSpatialGraph(sce, img_id = "ImageNb",
type = "knn",
name = "knn_sc_graph",
k = 15)
#> The returned object is ordered by the 'ImageNb' entry.
sce <- aggregateNeighbors(sce, colPairName = "knn_sc_graph",
aggregate_by = "metadata",
count_by = "cellular_neighborhood",
name = "aggregatedNeighborhood")
# Detect spatial context
sce <- detectSpatialContext(sce, entry = "aggregatedNeighborhood",
threshold = 0.9)
plotSpatial(sce, img_id = "ImageNb",
colPairName = "knn_sc_graph",
node_color_by = "spatial_context",
scales = "free")
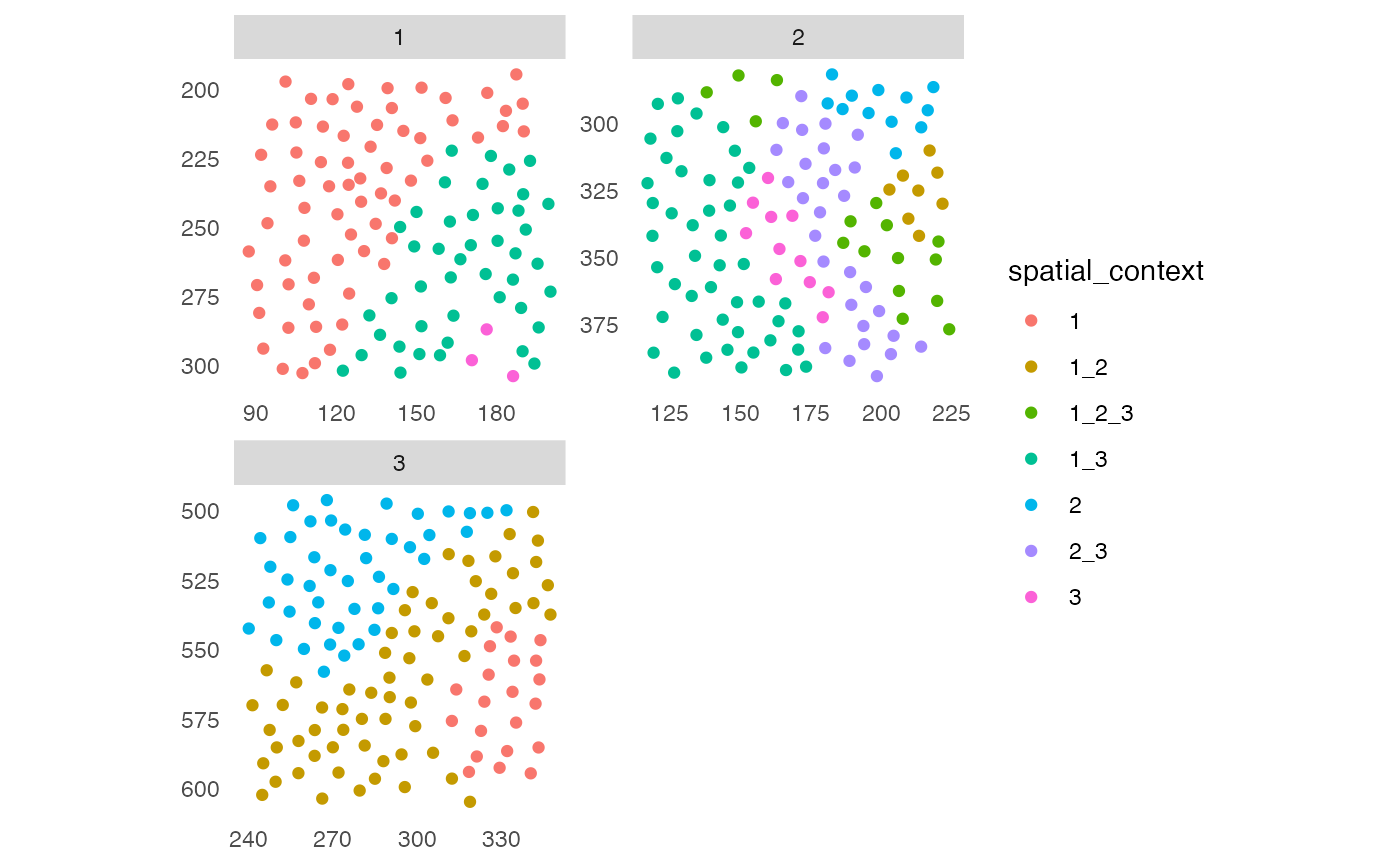 # Plot spatial context - default
plotSpatialContext(sce, group_by = "ImageNb")
# Plot spatial context - default
plotSpatialContext(sce, group_by = "ImageNb")
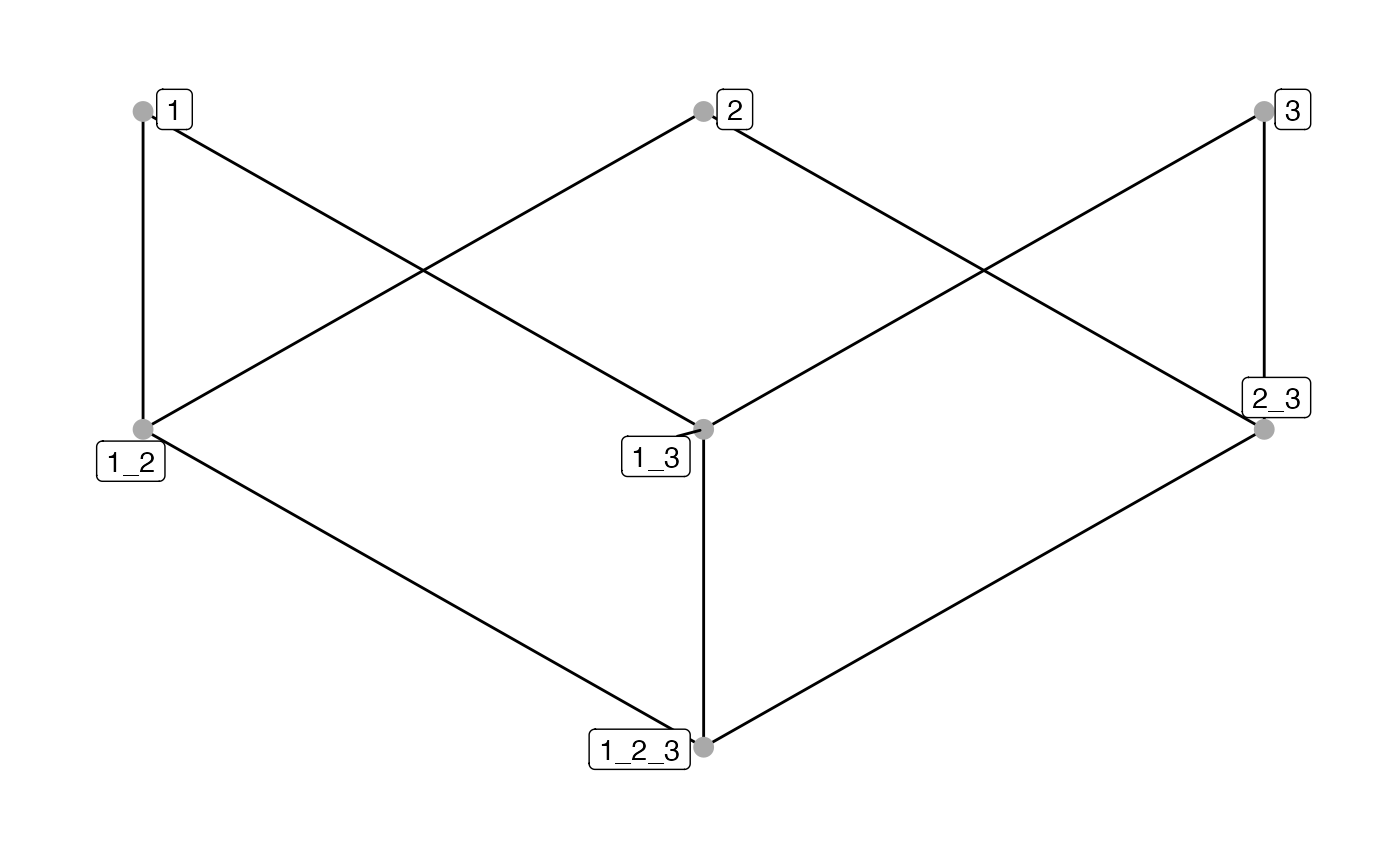 # Plot spatial context - adjust aesthetics
plotSpatialContext(sce, group_by = "ImageNb",
node_color_by = "name",
node_size_by = "n_cells",
node_label_color_by = "name")
# Plot spatial context - adjust aesthetics
plotSpatialContext(sce, group_by = "ImageNb",
node_color_by = "name",
node_size_by = "n_cells",
node_label_color_by = "name")
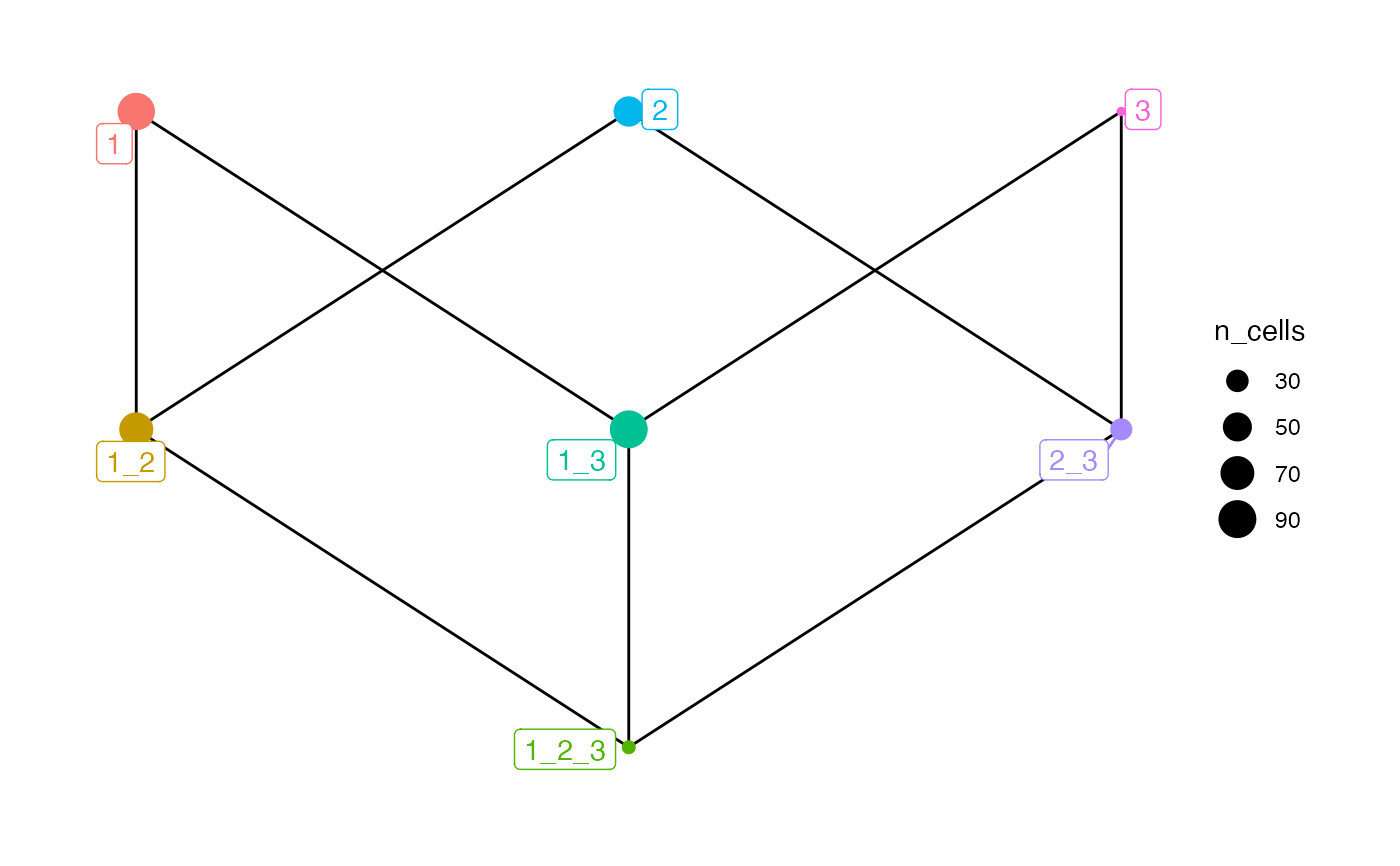 plotSpatialContext(sce, group_by = "ImageNb",
node_color_by = "n_cells",
node_size_by = "n_group")
plotSpatialContext(sce, group_by = "ImageNb",
node_color_by = "n_cells",
node_size_by = "n_group")
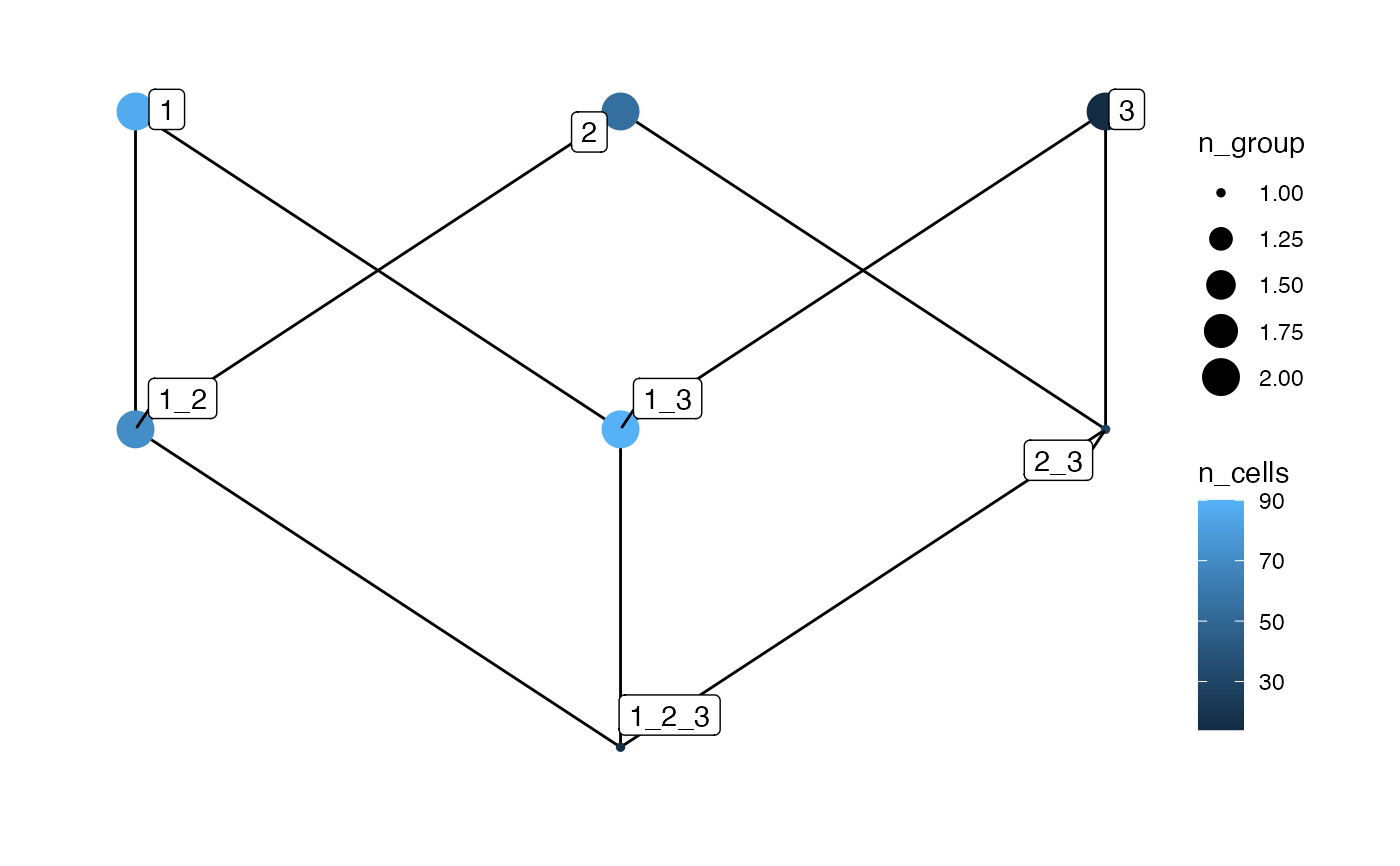 # Plot spatial context - return data
plotSpatialContext(sce, group_by = "ImageNb",
return_data = TRUE)
#> $edges
#> from to
#> 1 1 1_2
#> 2 1 1_3
#> 3 1_2 1_2_3
#> 4 1_3 1_2_3
#> 5 2 1_2
#> 6 2 2_3
#> 7 2_3 1_2_3
#> 8 3 1_3
#> 9 3 2_3
#>
#> $vertices
#> spatial_context n_cells n_group length
#> 1 1 87 2 1
#> 2 1_2 71 2 2
#> 3 1_2_3 16 1 3
#> 4 1_3 90 2 2
#> 5 2 55 2 1
#> 6 2_3 29 1 2
#> 7 3 14 2 1
#>
# Plot spatial context - return data
plotSpatialContext(sce, group_by = "ImageNb",
return_data = TRUE)
#> $edges
#> from to
#> 1 1 1_2
#> 2 1 1_3
#> 3 1_2 1_2_3
#> 4 1_3 1_2_3
#> 5 2 1_2
#> 6 2 2_3
#> 7 2_3 1_2_3
#> 8 3 1_3
#> 9 3 2_3
#>
#> $vertices
#> spatial_context n_cells n_group length
#> 1 1 87 2 1
#> 2 1_2 71 2 2
#> 3 1_2_3 16 1 3
#> 4 1_3 90 2 2
#> 5 2 55 2 1
#> 6 2_3 29 1 2
#> 7 3 14 2 1
#>